100% Spot Matching
Complete Expression Profiles with Delta2D
Delta2D's 100% spot matching produces complete expression profiles for every protein. Besides higher throughput, this leads to significantly improved statistical confidence, so you can, for example, identify more biomarker candidates from the same experiment. Other approaches to spot detection and matching lead to inconsistencies like missing values in expression profiles, and ambiguities in the profiles themselves. Delta2D's 100% spot matching is based on advanced image processing methods. It was introduced by DECODON in 2003.
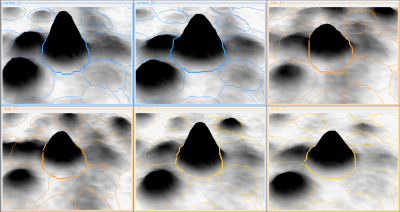
Here is a set of gels analyzed with Delta2D's 100% spot matching:
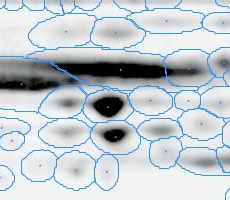
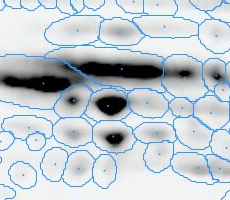
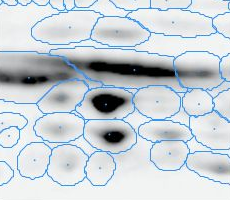
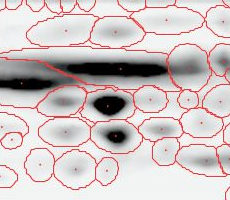
Note: The fourth image (with red spot boundaries) is a synthetic fusion image that is used in the process, see below.
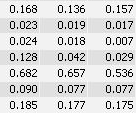
You see that the spot boundary patterns on each image are essentially the same. When a spot is not present on one of the gel images, Delta2D will still produce a segment there, with a quantity near zero. Notice that the expression profiles in the table have no gaps. This is comparable to the situation in transcriptomics: in a set of DNA arrays, you can always find corresponding spots across multiple arrays — they are at the same row and column on each array.
For reviewing a certain spot across all images Delta2D includes the Gel Image Regions view, which allows the detailled inspection of the same spot across all images even in a 3D perspective, e.g. if you are in doubt whether there is a spot or not or whether the spot should be split into two.
You can synchronously
- rotate a spot on all images,
- zoom in and out, and
- review a region even if there is no detected spot.
Furthermore you can select a certain spot which is visible in the 3D view:
The Benefits of Delta2D's 100% Spot Matching
Better statistical analysis
Complete expression profiles mean that more changes in spot patterns can be identified with statistical significance. Thus you can, for example, identify a larger set of biomarker candidates from the same raw data.
Increased productivity
Spot detection and editing is only done for the fusion image, as opposed to each image. For a 24 gel experiment this means you only have to check and correct one set of spot boundaries, as opposed to 24 different sets.
Better reproducibility
No hand-editing of spot matches means less potential for human error and operator-dependent variation. Furthermore, variation that is due to variations in the laboratory process, like sample preparation, or electrophoresis conditions etc. are much easier to detect when they are not obscured by matching problems.
Comparison to the Traditional Spot Matching Approach
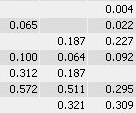
With traditional software packages, you have to edit the detected spots on each gel image, and resolve matching conflicts by inspecting all images involved. Here is an example of spot patterns and table of spot quantities produced by the traditional "separate detection on each gel image" approach (default parameters used, no spot editing).


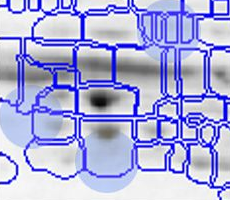
While you can correct many of the mismatches by hand, this process takes a lot of time and still does not resolve all ambiguities. The remaining spot matching ambiguities are resolved by the traditional software according to some black-box rules, producing frequent "missing values" in the expression profiles. A missing value can mean that there really was no spot on that gel, that there was a spot, but the software did not detect it, or that the software was unable to find the correct matching. In any case the expression profiles contain gaps that diminish the statistical confidence in subsequent quantitative analyses.
How Delta2D Achieves 100% Spot Matching
The benefits of 100% Spot Matching become true if you follow the recommended workflow.
